Introduction
Introduction
Structure comparison, as well as structure summarization, is a ubiquitous problem, appearing across multiple scientific disciplines. In particular, many scientific problems (e.g. inference of molecular properties from structure, pattern matching in data point clouds and scientific images) may be reduced to the problem of inexact graph matching: given two graphs, compute a measure of similarity that gainfully captures structural correspondence between the two. Similarly, many algorithms for addressing multiple scales of dynamical behavior rely on methods for automatically coarsening the computational graph associated with some model architecture.
In this work we present a distance metric for undirected graphs, based on the Laplacian exponential kernel. This measure generalizes the work of Hammond et al. (Hammond, Gur, and Johnson 2013) on graph diffusion distance for graphs of equal size; crucially, our distance measure allows for graphs of inequal size. We formulate the distance measure as the solution to an optimization problem dependent on a comparison of the two graph Laplacians. This problem is a nested optimization problem, with the innermost layer consisting of multivariate optimization subject to matrix constraints (e.g. orthogonality). To compute this dissimilarity score efficiently, we also develop and demonstrate the lower computational cost of an algorithm which calculates upper bounds on the distance. This algorithm finds a prolongation/restriction operator, , which produces an optimally coarsened version of the Laplacian matrix of a graph. Prolongation/restriction operators produced via the method in this paper can be used to accelerate the training of neural networks (both flat ANNs, as we will see in Chapter 6, and graph neural networks, as we will see in Chapter 7).
Prior Work
Quantitative measures of similarity or dissimilarity between graphs have been studied for decades owing to their relevance for problems in pattern recognition including structure-based recognition of extended and compound objects in computer vision, prediction of chemical similarity based on shared molecular structure, and many other domains. Related problems arise in quantitative modeling, for example in meshed discretizations of partial differential equations and more recently in trainable statistical models of data that feature graph-like models of connectivity such as Bayes Networks, Markov Random Fields, and artificial neural networks. A core problem is to define and compute how “similar” two graphs are in a way that is invariant to a permutation of the vertices of either graph, so that the answer doesn’t depend on an arbitrary numbering of the vertices. On the other hand unlike an arbitrary numbering, problem-derived semantic labels on graph vertices may express real aspects of a problem domain and may be fair game for detecting graph similarity (we explore the use of edge information in Section 8.2. The most difficult case occurs when such labels are absent, for example in an unstructured mesh, as we shall assume. Here we detail several measures of graph dissimilarity, chosen by historical significance and similarity to our measure.
We mention just a few prior works to give an overview of the development of graph distance measures over time, paying special attention to those which share theoretical or algorithmic characteristics with the measure we introduce. Our mathematical distinctions concern the following properties:
Does the distance measure require an inner optimization loop? If so is it mainly a discrete or continuous optimization formulation?
Does the distance measure calculation naturally yield some kind of explicit map from real-valued functions on vertices of one graph to functions on vertices of the other? (A map from vertices to vertices would be a special case.) If we use the term “graph signal” to mean a function which identifies each vertex of a graph with some state , then a map-explicit graph distance is one which as part of its output provides a new function which approximates the behavior of . ‘Approximates’ and ‘behavior’ are here left undefined as these would need to be problem-specific.
Is the distance metric definable on the spectrum of the graph alone, without regard to other data from the same graph? The “spectrum” of a graph is a graph invariant calculated as the eigenvalues of a matrix related to the adjacency matrix of the graph. Depending on context, the spectrum can refer to eigenvalues of the adjacency matrix, graph Laplacian, or normalized graph Laplacian of a graph. We will usually take the underlying matrix to be the graph Laplacian, defined in detail in Section 1.4.2. Alternatively, does it take into account more detailed “structural” aspects of the graph? This categorization (structural vs. spectral) is similar to that introduced in (Donnat and Holmes 2018).
For each of the graph distance variants discussed here, we label them according to the above taxonomy. For example, the two prior works by Eschera et. al. and Hammond et al (discussed in Sections 1.2.4 and 1.2.5) would be labelled as (structural, explicit, disc-opt) and (spectral, implicit, non-opt), respectively. Our distance measure1 defined in detail in Chapter 2 would be labelled (spectral, explicit, cont-opt).
Quadratic Matching of Points and Graphs (structural, explicit, cont-opt)
As a first example, some graph comparison methods focus on the construction of a point-to-point correspondence between the vertices of two graphs. Gold et. al. (Gold et al. 1995) define the dissimilarity between two unlabelled weighted graphs (with adjacency matrices and and and vertices, respectively) as the solution to the following optimization problem (for real-valued :
$$\label{eqn:gold_defn} \begin{array}{ll@{}ll} \text{minimize} & \displaystyle \sum\limits_{j=1}^{n_2}\sum\limits_{k=1}^{n_1} {\left( \sum\limits_{l=1}^{n_2} A^{(1)}_{jl} m_{lk} - \sum\limits_{p=1}^{n_1} m_{j p} A^{(2)}_{pk} \right)}^2 &= {\left|\left| A^{(1)} M - M A^{(2)} \right| \right|}_F^2\\ \text{subject to}& \displaystyle\sum\limits_{i=1}^{n_2} m_{ij} = 1, &j=1 \ldots n_1\\ & \displaystyle\sum\limits_{j=1}^{n_1} m_{ij} = 1, &i=1 \ldots n_2\\ & m_{ij} \geq 0 & i = 1 \ldots n_2\\ & &j = 1 \ldots n_1 \\ \end{array}$$ where is the squared Frobenius norm. This problem is similar in structure to the optimization considered in Section 2.4 and Chapter 4: a key difference being that Gold et al. consider optimization over real-valued matchings between graph vertices, whereas we consider 0-1 valued matchings between the eigenvalues of the graph Laplacians. In (Gold, Rangarajan, and Mjolsness 1995) and (Rangarajan, Gold, and Mjolsness 1996) the authors present computational methods for computing the optimum of [eqn:gold_defn], and demonstrate applications of this distance measure to various machine learning tasks such as 2D and 3D point matching, as well as graph clustering. Gold et al. also introduce softassign, a method for performing combinatorial optimization with both row and column constraints, similar to those we consider.
Cut-Distance of Graphs (structural, implicit, disc-opt)
Lovász (Lovász 2012) defines the cut-distance of a pair of graphs as follows: Let the -norm of a matrix be given by:
Given two labelled graphs , , on the same set of vertices, and their adjacency matrices and , the cut-distance is then given by (for more details, see (Lovász 2012)). Computing this distance requires combinatorial optimization (over all vertex subsets of ) but this optimization does not result in an explicit map between and . This distance metric is grounded in the theory of graphons, mathematical objects which are a natural infinite-sized generalization of dense graphs. However, all sparse graphs are similar in cut-distance to the zero graphon (see (Lovász 2012)), making cut-distance less useful for real-world problems.
Wasserstein Earth Mover Distance (spectral, implicit, disc-opt)
One common metric between graph spectra is the Wasserstein Earth Mover Distance. Most generally, this distance measures the cost of transforming one probability density function into another by moving mass under the curve. If we consider the eigenvalues of a (possibly weighted) graph as point masses, then the EMD measures the distance between the two spectra as the solution to a transport problem (transporting one set of points to the other, subject to constraints e.g. a limit on total distance travelled or a limit on the number of ‘agents’ moving points). The EMD has been used in the past in various graph clustering and pattern recognition contexts; see (Gu, Hua, and Liu 2015). In the above categorization, this is an optimization-based spectral distance measure, but is implicit, since it does not produce a map from vertices of to those of (informally, this is because the EMD is not translating one set of eigenvalues into the other, but instead transforming their respective histograms). Recent work applying the EMD to graph classification includes (Dodonova et al. 2016) and (McGregor and Stubbs 2013). Some similar recent works (Maretic et al. 2019; Chen et al. 2020) have used optimal transport theory to compare graphs. In this framework, signals on each graph are smoothed, and considered as draws from probability distribution(s) over the set of all graph signals. An optimal transport algorithm is used to find the optimal mapping between the two probability distributions, thereby comparing the two underlying graphs.
Graph-Edit Distance
The graph edit distance measures the total cost of converting one graph into another with a sequence of local edit moves, with each type of move (for example, vertex deletion or addition, edge deletion or addition, edge division or contraction) incurring a specified cost. Costs are chosen to suit the graph analysis problem at hand; determining a cost assignment which makes the edit distance most instructive for a certain set of graphs is both problem-dependent and an active area of research. The distance measure is then the sum of these costs over an optimal sequence of edits, which must be found using some optimization algorithm i.e. a shortest-path algorithm (the best choice of algorithm may vary, depending on how the costs are chosen). The sequence of edits may or may not (depending on the exact set of allowable edit moves) be adaptable into an explicit map between vertex-sets. Classic pattern recognition literature includes: (Eshera and Fu 1984) (Eshera and Fu 1986) (Gao et al. 2010) (Sanfeliu and Fu 1983) .
Diffusion Distance due to Hammond et al. (Hammond, Gur, and Johnson 2013)
We discuss this recent distance metric more thoroughly below. This distance measures the difference between two graphs as the maximum discrepancy between probability distributions which represent single-particle diffusion beginning from each of the nodes of and . This distance is computed by comparing the eigenvalues of the heat kernels of the two graphs. The optimization involved in calculating this distance is a simple unimodal optimization over a single scalar, , representing the passage of time for the diffusion process on the two graphs; hence we do not count this among the “optimization based” methods we consider.
Novel Diffusion-Derived Measures
In this work, we introduce a family of related graph distance measures. These measures compare two graphs in terms of similarity of a set of probability distributions describing single-particle diffusion on each graph. For two graphs and with respective Laplacians and , the matrices and are called the Laplacian Exponential Kernels of and ( is a scalar representing the passage of time). The column vectors of these matrices describe the probability distribution of a single-particle diffusion process starting from each vertex, after time has passed. The norm of the difference of these two kernels thus describes how different these two graphs are, from the perspective of single-particle diffusion, at time . Since these distributions are identical at very-early and very late times (we formalize this notion in Section 2.1), a natural way to define a graph distance is to take the supremum over all 2. When the two graphs are the same size, so are the two kernels, which may therefore be directly compared with a matrix norm. This case is the case considered by Hammond et al. (Hammond, Gur, and Johnson 2013). However, to compare two graphs of different sizes, we need a mapping between the column vectors of and .
One such mapping is optimization over a suitably constrained prolongation/restriction operator between the graph Laplacians of the two graphs. This operator is a permutation-invariant way to compare the behavior of a diffusion process on each. The prolongation map thus calculated may then be used to map signals (by which we mean values associated with vertices or edges of a graph) on to the space of signals on (and vice versa). In Chapters 6 and 7 we implicitly consider the weights of an artificial neural network model to be graph signals, and use these operators to train a hierarchy of linked neural network models.
We also, in sections 3.3 and 3.4 consider a time conversion factor between diffusion on graphs of unequal size, and consider the effect of limiting this optimization to sparse maps between the two graphs (again, our case reduces to Hammond when the graphs in question are the same size, dense and matrices are allowed, and our time-scaling parameter is set to 1).
In this work, we present an algorithm for computing the type of nested optimization given in our definition of distance (Equations [defn:exp_defn] and [defn:linear_defn]). The innermost loop of our distance measure optimization consists of a Linear Assignment Problem (LAP, defined below) where the entries of the cost matrix have a nonlinear dependence on some external variable. Our algorithm greatly reduces both the count and size of calls to the external LAP solver. We use this algorithm to compute an upper bound on our distance measure, but it could also be useful in other similar nested optimization contexts: specifically, nested optimization where the inner loop consists of a linear assignment problem whose costs depend quadratically on the parameter in the outermost loop.
Outline
The goal of this manuscript is to develop the theory and practice of comparing graphs using Graph Diffusion Distance (GDD). The remainder of this chapter (Chapter 1) defines basic mathematical terminology and framework necessary for the remainder of the work. Chapter 2 defines Graph Diffusion Distance and the variants thereof considered. Efficiently computing these distance metrics requires a novel algorithm, which we motivate and explain in Chapter 4. Chapters 3 and 5 explore theoretical and numeric properties of GDD, respectively. Chapters 6, 7, and 8 showcase several applications of GDD to various scientific tasks. Chapters 6 and 7 in particular are structured as self-contained investigations and may be read without material from Chapters 2-5, although material from Section 1.4 may be necessary for understanding notation.
Mathematical Background
In this section we briefly define terminology and notation which will be useful in the exposition and proofs to follow.
Desirable Characteristics for Distance Metrics
The ideal for a quantitative measure of similarity or distance on some set is usually taken to be a distance metric satisfying for all :
Non-negativity:
Identity:
Symmetry:
Triangle inequality:
Then is a metric space. Euclidean distance on and geodesic distance on manifolds satisfy these axioms. They can be used to define algorithms that generalize from to other spaces. A variety of weakenings of these axioms are required in many applications, by dropping some axioms and/or weakening others. For example if is a set of nonempty sets of a metric space , one can define the “Hausdorff distance” on which is an extended pseudometric that obeys the triangle inequality but not the Identity axiom and that can take values including . As another example, any measure measure of distance on graphs which is purely spectral (in the taxonomy of Section 1.2) cannot distinguish between graphs which have identical spectra. We discuss this in more detail in Section 2.3.
Additional properties of distance metrics that generalize Euclidean distance may pertain to metric spaces related by Cartesian product, for example, by summing the squares of the distance metrics on the factor spaces. We will consider an analog of this property in Section 3.6.
Definitions
[subsub:definitions]
Graph Laplacian: For an undirected graph with adjacency matrix and vertex degrees , we define the Laplacian of the graph as The eigenvalues of this matrix are referred to as the spectrum of . See (Belkin and Niyogi 2002; Cvetkovic, Rowlinson, and Simic 2010) for more details on graph Laplacians and spectral graph theory. is sometimes instead defined as ; we take this sign convention for because it agrees with the standard continuum Laplacian operator, , of a multivariate function : .
Frobenius Norm: The squared Frobenius norm, of a matrix is given by the sum of squares of matrix entries. This can equivalently be written as $\Tr[A^T A]$.
Linear Assignment Problem (LAP): We take the usual definition of the Linear Assignment Problem (see (Burkard and Cela 1999), (Burkard, Dell’Amico, and Martello 2009)): we have two lists of items and (sometimes referred to as “workers” and “jobs”), and a cost function which maps pairs of elements from and to an associated cost value. This can be written as a linear program for real-valued as follows: $$\label{eqn:lap_defn} \begin{array}{ll@{}ll} \text{minimize} & \displaystyle\sum\limits_{i=1}^{m}\sum\limits_{j=1}^{n} c(s_i,r_j)x_{ij} &\\ \text{subject to}& \displaystyle\sum\limits_{i=1}^{m} x_{ij} \leq 1, &j=1 \ldots n\\ & \displaystyle\sum\limits_{j=1}^{n} x_{ij} \leq 1, &i=1 \ldots m\\ & x_{ij} \geq 0 & i = 1 \ldots m, j = 1 \ldots n \\ \end{array}$$ Generally, “Linear Assignment Problem” refers to the square version of the problem where , and the objective is to allocate the jobs to workers such that each worker has exactly one job and vice versa. The case where there are more workers than jobs, or vice versa, is referred to as a Rectangular LAP or RLAP. In practice, the conceptually simplest method for solving an RLAP is to convert it to a LAP by augmenting the cost matrix with several columns (rows) of zeros. In this case, solving the RLAP is equivalent to solving a LAP with size . Other computational shortcuts exist; see (Bijsterbosch and Volgenant 2010) for details. Since the code we use to solve RLAPs takes the augmented cost matrix approach, we do not consider other methods in this paper.
Matching: we refer to a 0-1 matrix which is the solution of a particular LAP as a “matching”. We may refer to the “pairs” or “points” of a matching, by which we mean the pairs of indices with . We may also say in this case that “assigns” to . Given two matrices and , and lists of their eigenvalues and , with , we define the minimal eigenvalue matching as the matrix which is the solution of the following constrained optimization problem: In the case of eigenvalues with multiplicity , there may not be one unique such matrix, in which case we distinguish matrices with identical cost by the lexicographical ordering of their occupied indices and take as the first of those with minimal cost. This matching problem is well-studied and efficient algorithms for solving it exist; we use a Python language implementation (Clapper, n.d.) of a 1957 algorithm due to Munkres (Munkres 1957). Additionally, given a way to enumerate the minimal-cost matchings found as solutions to this eigenvalue matching problem, we can perform combinatorial optimization with respect to some other objective function , in order to find optima of subject to the constraint that is a minimal matching.
Hierarchical Graph Sequences: A Hierarchical Graph Sequence (HGS) is a sequence of graphs, indexed by , satisfying the following:
is the graph with one vertex and one self-loop, and;
Successive members of the lineage grow roughly exponentially - that is, there exists some base such that the growth rate (of nodes) as a function of level number is , for all .
Graded Graph: A graded graph is a graph along with a vertex labelling, where vertices are labelled with non-negative integers such that , the difference in label over any edge, is in . We will refer to the edges as “within-level” and the edges as “between-level”.
Graph Lineages: A graph lineage is a graded graph with two extra conditions:
The vertices and edges with form a HGS; and
the vertices and edges with form a HGS of bipartite graphs.
More plainly, a graph lineage is an exponentially growing sequence of graphs along with ancestry relationships between nodes. We will also use the term graph lineage to refer to the HGS in the first part of the definition (it will be clear from context which sense we are using). Some intuitive examples of graph lineages in this latter sense are the following:
Path graphs or cycle graphs of size for any integer .
More generally, grid graphs of any dimension , of side length , yielding a lineage which grows with size (with periodic or nonperiodic boundary conditions).
For any probability distribution whose support is points in the unit square, we can construct a graph by discretizing the map of as a function of and , and interpreting the resulting matrix as the adjacency matrix of a graph. For a specific probability distribution , the graphs derived this way with discretizations of exponentially increasing bin count form a graph lineage.
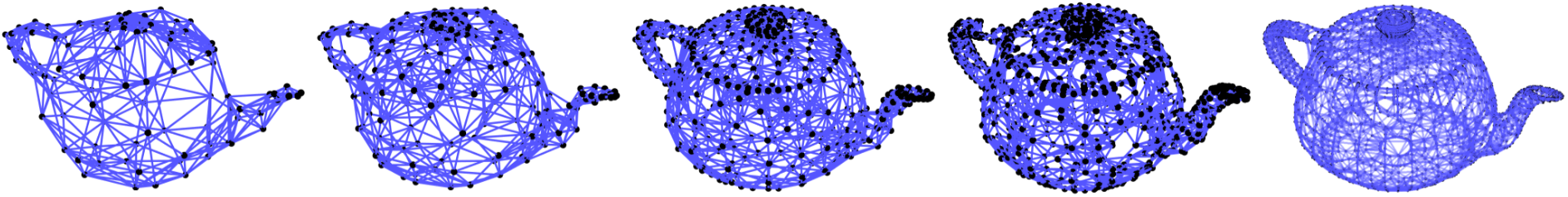
The triangulated mesh is a common object in computer graphics (Patney, Ebeida, and Owens 2009; Morvan and Thibert 2002; Shamir 2008), representing a discretization of a 2-manifold embedded in . Finer and finer subdivisions of such a mesh constitute a graph lineage.
Several examples of graph lineages are used in the discussion of the numerical properties of Graph Diffusion Distance in Section 5.1. Additional examples (a path graph and a triangulated mesh) can be found in Figures [fig:graph_lineages_1] and [fig:graph_lineages_2].
[fig:graph_lineages_1]
[fig:graph_lineages_2]
Kronecker Product and Sum of matrices: Given a matrix , and some other matrix , the Kronecker product is the block matrix See (Hogben 2006), Section 11.4, for more details about the Kronecker Product. If and are square, their Kronecker Sum is defined, and is given by where we write to denote an identity matrix of the same size as .
Box Product () of graphs: For with vertex set and with vertex set , is the graph with vertex set and an edge between and when either of the following is true:
and and are adjacent in , or
and and are adjacent in .
This may be rephrased in terms of the Kronecker Sum of the two matrices: Cross Product () of graphs: For with vertex set and with vertex set , is the graph with vertex set and an edge between and when both of the following are true:
and are adjacent in , and
and are adjacent in .
We include the standard pictorial illustration of the difference between these two graph products in Figure [fig:graph_prod].

[fig:graph_prod]
Grid Graph: a grid graph (called a lattice graph or Hamming Graph in some texts (Brouwer and Haemers 2012)) is the distance-regular graph given by the box product of path graphs (yielding a grid with aperiodic boundary conditions) or by a similar list of cycle graphs (yielding a grid with periodic boundary conditions).
Prolongation map: A prolongation map between two graphs and of sizes and , with , is an matrix of real numbers which is an optimum of the objective function of equation [eqn:objfunction] below (possibly subject to some set of constraints ).