The overarching goal of this project is to develop analytic methods that enable scientists to efficiently and automatcially discover relationships between shape and function in biological systems. This work has been supported by the NSF Emerging Frontiers in the form of a large collaborative effort on bioshape (bioshapes.org, DBI-1053036) and a collaborative project supported by NSF Division of Biological Infrastructure Division on bioimage analysis for understanding pollen morphology and texture (DBI-1262547). Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation.
Pollen Morphology-based Classification and Analysis (with Punyasena Lab at UIUC)
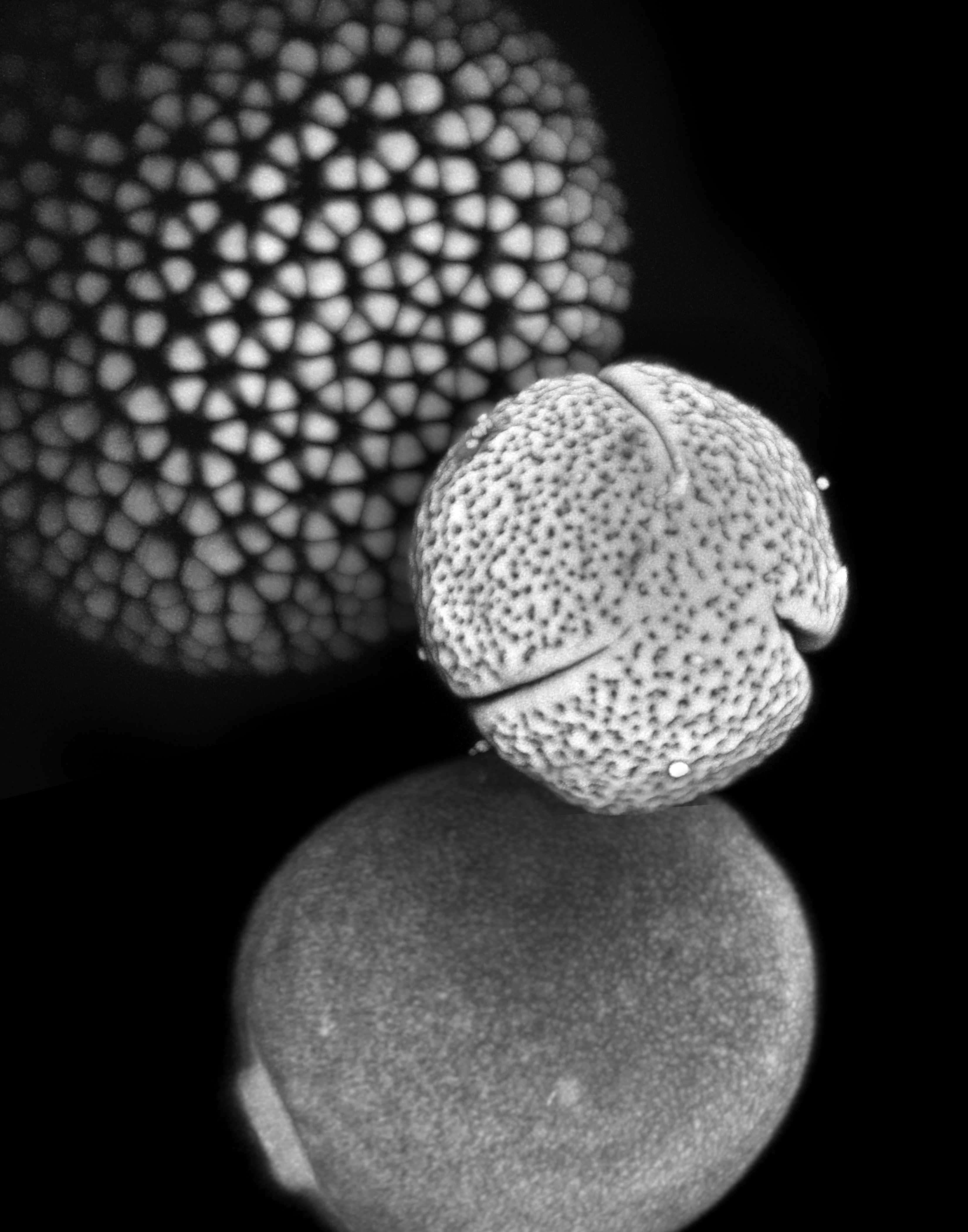
The practice of identifying pollen has a large number of scientific applications and is used in fields as diverse as archaeology, biostratigraphy (the dating of rocks), and forensic science. Pollen and spores play a particularly important role in paleontology, because they form the most abundant and extensive record of plant diversity, dating back hundreds of millions of years. However, many critical hypotheses in plant ecology and evolution (e.g. the assembly of plant communities, speciation and extinction) cannot be fully tested with pollen data due to the extreme difficulty of recognizing species from pollen and spore material.
We are developing methods for automatically classifying pollen based on texture and shape features. This approach has been validated in distinguishing visually similar SEM images of pollen from several different species based on fine scale textural differences, recognition of fossil pollen grains based on training from modern grains, and automatic detection, segmentation and classification of pollen grains in wide-field imagery.
Related Publications:
- S. Kong, C. Fowlkes, "Low-rank Bilinear Pooling for Fine-Grained Classification",
CVPR, (July 2017).
arXiv:1611.05109
[pdf]
- B. Kong, C. Fowlkes,
"Energy-Based Spherical Sparse Coding", Technical Report (2017),
arXiv:1710.01820
[pdf]
-
G. Ghiasi, C. Fowlkes, "Laplacian Pyramid Reconstruction and Refinement for Semantic Segmentation",
ECCV, Amsterdam, (Oct. 2016).
arXiv:1605.02264
[pdf]
[code]
-
S. Kong, S. Punyasena, C. Fowlkes, "Spatially Aware Dictionary Learning and Coding for Fossil Pollen Identiifcation",
CVMI Workshop, Los Vegas, NV, (July 2016).
arXiv:1605.00775
[pdf]
- D. Tcheng, A. Nayak, C. Fowlkes, S. Punyasena,
"Visual recognition software for binary classification and its application to spruce pollen identification"
PLoS ONE,
11(2): e0148879. doi:10.1371/journal.pone.0148879.
[pdf]
- L. Mander, M. Li, W. Mio, C. Fowlkes, S. Punyasena, "Classification
of grass pollen through the quantitative analysis of surface ornamentation and
texture",
Proc. R. Soc. B. 2013 280 (1770)
[pdf]
- B. Kong, C. Fowlkes, "Fast Convolutional Sparse Coding", UCI
Technical Report, May 2014
[pdf]
J. Yarkony, C. Fowlkes, "Planar Ultrametrics for Image Segmentation", Proc. of , NIPS, Dec. 2015. arXiv:1507.02407 [pdf]
- S. Punyasena, L. Mander, M. Li, W. Mio, C. Fowlkes, Classifying grass
pollen using high-resolution imaging and the quantitative analysis of surface
texture, Botany and Plant Biology 2013 Joint Congress. Botanical Society of
America,New Orleans, LA, July 2013
- S. Punyasena, D. Tcheng, C. Fowlkes, W. Mio, C-R. Shyu.
Digital palynology: imaging, automation and intelligent databases. Abstracts
with Programs – Geological Society of America, Geological Society of America,
2014 annual meeting, Vancouver, Canada, Oct. 2014
- D. Haselhorst, S. Kong, C. Fowlkes, J. Moreno, D. Tcheng, S. Punyasena,
``Automating tropical pollen counts using convolutional neural nets: from image
acquisition to identification'' Inaugural Digital Data in Biodiversity Research
Conference, Ann Arbor, MI, June 2017
- M. Urban, M. Sivaguru, I. Romero, G. Fried, C. Fowlkes, W. Mio, C.
Jaramillo, and S. Punyasena, ``The application of optical super-resolution
microscopy to the study of pollen morphology''. Inaugural Digital Data in
Biodiversity Research Conference, Ann Arbor, MI, June 2017.
- I. Romero, S. Kong, C. Fowlkes, M. Urban, C. D'Apolito, C. Jaramillo, F.
Oboh-Ikuenobe, S. Punyasena ``Cenozoic biogeography of Striatopollis catatumbus
(Fabaceae Detariae) as a case of study of the evolution of the Neotropical
flora'', 50th Annual Meeting of AASP - The Palynological Society, Nottingham,
UK, Sept. 2017
- I. Romero, S. Kong, C. Fowlkes, M. Urban, C. D'Apolito, C. Jaramillo, F.
Oboh-Ikuenobe, S. Punyasena ``Novel Morphological Analysis of a Fossil Fabaceae
Pollen Type, Striatopollis Catatumbus (Tribe Detariae)'', Geological Society of
America Annual Meeting, Seattle Washington, Oct. 2017
Software Tools and Demos:
- Texture recognition using sparse coding histograms and nearest-neighbor classification:
This code include a MATLAB demo for classifying grass pollen SEM images
(dataset from Mander et al 2013) which achieves ~77 percent accuracy in
species identification.
Sparse coding for texture classification:[texture_coding_0.2.tar.gz]
- Fast convolutional sparse coding:
An implementation by Bailey Kong
as described in our tech report.
This provides a mechanism for effeciently learning a filter dictionary
for encoding pollen surface texture.
Fast convolutional sparse coding: [code on github]
-
Spatially-aware Dictionary Learning and Coding for Pollen Identification.
This code (implemented by Shu
Kong) performs spatially-aware dictionary learning for matching
surface texture of spruce pollen grains. The method and results are
described in Kong et. al.
2016. There is also a separate code repository here
that describes the algorithm used for selecting patches based on greedy
submodular optimization.
Spatially-aware Dictionary Learning: [code on github]
Identifying fossil pollen from modern reference: [code on github]
-
Joint Pollen Grain Detection and Classification with Multiplicative Gating Architecture:
This code implements joint segmentation and classification of pollen grains in wide-field
transmitted light images using multiplicative feature masking for the
classification step. Implementation by Shu Kong.
Joint Detection and Classification: [code on github]
-
Automated recognition and counting of pollen grain species from field samples:
This code implements an (unpublished) pipeline for detecting,
segmenting and classifying pollen grains in wide-field transmitted
light images. Implementation by Shu Kong.
Automated pollen species abundance pipeline: [code on github]
 Adaptive software for automated detection, tracking and segmentation
(with Cinquin Lab at UCI)
Adaptive software for automated detection, tracking and segmentation
(with Cinquin Lab at UCI)While the particular uses of images as an experimental tool in biology vary widely, the common tasks of recognition, grouping and tracking are ubiquitous. At present, automated image analysis is typically limited to large scale projects that can justify the resources required for development of custom software. The resulting systems are often brittle and require redevelopment each time a slightly different dataset appears. We are working on generic tools that can be interactively retrained by the end user with the goal of expanding the role of quantitative imaging and modeling in biology by “mining the long tail” of under-analyzed experimental data.
Publications:
- A. Cinquin, M. Chiang, A. Paz, S. Hallman, O. Yuan, C. Fowlkes, O.
Cinquin, "Intermittent stem cell cycling balances self-renewal and senescence
of the C. elegans germ line", in press, PLoS Genetics (2016)
- M. Chiang, S. Hallman, A. Cinquin, N. Reyes de Mochel, A. Paz, S. Kawauchi, A. Calof, K. Cho, C. Fowlkes, O. Cinquin,
"Analysis of in vivo single cell behavior by high throughput, human-in-the-loop segmentation of three-dimensional images",
BMC Bioinformatics 2015, 16:397 doi:10.1186/s12859-015-0814-7.
[pdf]
- S. Hallman, C. Fowlkes, "Oriented Edge Forests for Boundary Detection",
CVPR, Boston, MA, (June 2015).
arXiv:1412.2066 [pdf] [code]
-
X. Zhu, C. Vondrick, C. Fowlkes, D. Ramanan, "Do we need more training data?",
IJCV, DOI 10.1007/s11263-015-0812-2, 2015
[arXiv:1503.01508]
[pdf]
- S. Wang, C. Fowlkes, "Learning Optimal Parameters for Multi-target Tracking",
BMVC 2015
[pdf]
-
S. Hallman, C. Fowlkes, "Oriented Edge Forests for Boundary Detection",
CVPR, Boston, MA, (June 2015).
arXiv:1412.4181
[pdf]
[code]
Software:
- MATLAB code for training template based detectors for detecting cell nuclei in fluorescence image stacks.
- MATLAB code for learning and inference in multi-target tracking with pairwise interactions between objects.
Gene expression and developmental patterning in Drosophila (with DePace Lab at Harvard)
 For a gene to function properly, it must be active in the right place, at the
right time, and in the right amount. Changes in any of these features can lead
to observable differences between individuals and species and in some cases can
lead to disease. We do not currently understand how the position, timing, and
amount of gene expression is encoded in DNA sequence. One approach to this
problem is to compare how gene expression differs between species and to try to
relate changes in DNA sequence to changes in gene expression.
We are developing methods for measuring and comparing gene expression patterns
at high spatial and temporal resolution between embryos of different species of
Drosophila. The methods allow us to control for variation in the size, shape,
and number of nuclei between embryos.
For a gene to function properly, it must be active in the right place, at the
right time, and in the right amount. Changes in any of these features can lead
to observable differences between individuals and species and in some cases can
lead to disease. We do not currently understand how the position, timing, and
amount of gene expression is encoded in DNA sequence. One approach to this
problem is to compare how gene expression differs between species and to try to
relate changes in DNA sequence to changes in gene expression.
We are developing methods for measuring and comparing gene expression patterns
at high spatial and temporal resolution between embryos of different species of
Drosophila. The methods allow us to control for variation in the size, shape,
and number of nuclei between embryos.
Publications:
-
M. Staller, C. Fowlkes, M. Bragdon, J. Estrada, Z. Wunderlich, A. DePace, "A
gene expression atlas of a bicoid-depleted Drosophila embryo reveals early canalization of cell fate",
Development 142, p. 587-596, (2015)
[pdf]
- C. Fowlkes, K. Eckenrode, M. Bragdon, M. Meyer, Z. Wunderlich, L.
Simirenko, C. Hendriks, S. Keranen, C. Henreiquez, M. Biggin, M. Eisen,
A. DePace, "A conserved developmental patterning network produces
quantitatively different output in multiple species of Drosophila,"
PLoS Genetics,
7(10): e1002346, 2011. [pdf]
- C. Fowlkes, C. Luengo Hendriks, S. Keränen, G. Weber, O.
Rübel, M-Y Huang, S. Chatoor, L. Simirenko, A. DePace C.
Henriquez, A. Beaton, R. Weiszmann, S. Celniker, B. Hamann, D.
Knowles, M. Biggin, M. Eisen, J. Malik.
"Constructing a quantitative spatio-temporal atlas of gene expression in the Drosophila blastoderm",
Cell, 133(2), p. 364-374, 2008.
[pdf]
[supplement]
Software:
- PointCloudAlign toolkit (v1.0.1) for registering gene expression patterns acquired using the BDTNP PointCloudToolbox image processing pipeline
Quantification of cellular and sub-cellular alignment (with Khine Lab at UCI)
Nano- and microscale topographical cues play critical roles in the induction and maintenance of various cellular functions, including morphology, adhesion, gene regulation, and communication. Recent studies indicate that structure and function at the heart tissue level is exquisitely sensitive to mechanical cues at the nano-scale as well as at the microscale level. We have developed image analysis tools coupled with an inexpensive culture platform comprised of biomimetic wrinkles that simulate the heart’s complex anisotropic and multiscale architecture for facile and robust cardiac cell alignment. We demonstrate the cellular and subcellular alignment of both neonatal mouse cardiomyocytes as well as those derived from human embryonic stem cells. By mimicking the fibrillar network of the extracellular matrix, this system enables monitoring of protein localization in real time and therefore the high-resolution study of phenotypic and physiologic responses to in-vivo like topographical cues.
Publications:
- C. McCusker, A. Athippozhy, C. Diaz-Castillo, C. Fowlkes, D.
Gardiner, S. Voss, "Positional Plasticity in Regenerating Amybstoma
mexicanum Limbs Is Associated With Cell Proliferation and Pathways of
Cellular Differentiation", BMC Developmental Biology, 15:45,
DOI 10.1186/s12861-015-0095-4 (2015).
[pdf]
- A. Chen, D. Lieu, L. Freschauf, V. Lew, H. Sharma, J. Wang, D. Nguyen, I. Karakikes, R. Hajjar, A. Gopinathan, E.
Botvinick, C. Fowlkes, R. Li, M. Khine, "Shrink-Film Configurable Multiscale
Wrinkles for Functional Alignment of Human Embryonic Stem Cells and their
Cardiac Derivatives", 10.1002/adma.201103463, Advanced Materials, 2011.
[pdf]
- J. Luna, J. Ciriza, M. Ojeda-Garcia, M. Kong, A. Herren, D. Lieu, R. Li, C. Fowlkes, M. Khine, K. McCloskey,
"Multi-scale Biomimetic Topography for the Alignment of Neonatal and
Embryonic Stem Cell-derived Heart Cells",
Tissue Engineering: Part C , 17(5), p. 579-588 , 2011
[pdf]
- A. Chen, E. Lee, R. Tu, K. Santiago, A. Grosberg, C. Fowlkes, M. Khine, "Integrated Platform for Functional Monitoring of Biomimetic Heart Sheets Derived From Human Pluripotent Stem Cells", Biomaterials, 35(2):675-683. [pdf]
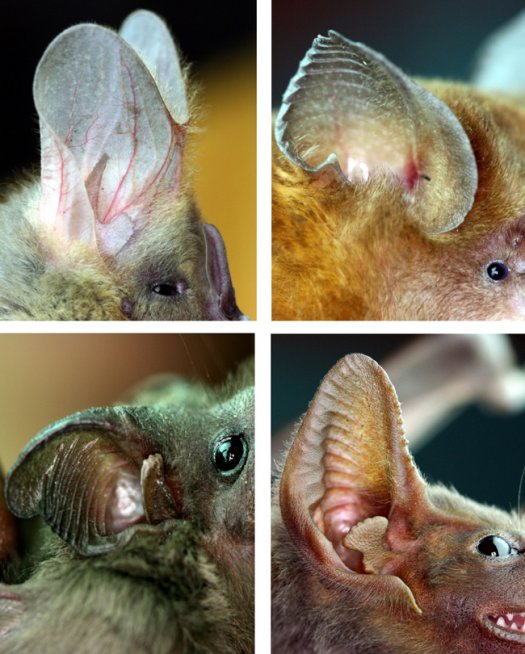
Functionally derived shape metrics for analyzing Bat Biosonar (with Mueller Lab at VT)
Statistical analysis of collections of shapes typically relies on some measure of distance between shapes which is specified a priori without consideration of the relevance of particular modes of variation to biological function. We are developing methods that leverage accurate simulation of acoustical propagation to understand the functional importance of aspects of bat ear and noseleaf shape. We exploit the adjoint method to efficiently perform a global analysis of the sensitivity of acoustical performance to shape features.
